The KEYNOTE-427 trial is a Phase II trial of first-line pembrolizumab in patients with clear cell (cohort A) or non-clear cell (cohort B) renal cell carcinoma. In cohort A (n=110 patients) pembrolizumab continued to demonstrate antitumor activity after a median follow up of 22.6 months, with an acceptable safety profile.
In this presented poster, the authors analyzed RNA sequencing-based signatures in patients with clear cell renal cell carcinoma treated with pembrolizumab monotherapy in the cohort A of the KEYNOTE-427 study.
The objective of the study was to evaluate the association between baseline RNA sequencing-based signatures and response or resistance to the pembrolizumab drug in patients with advanced clear cell renal carcinoma. The authors evaluated the association of 11 signatures quantifying the tumor microenvironment with clinical outcomes. Due to the fact that the statistical power was potentially limited, the authors decided that the significance level needs to be prespecified at the level of 0.1.
The baseline characteristics of the analyzed patients are shown in table 1. T-cell inflamed GEP was statistically significantly associated with an objective response rate (p=0.021) and overall survival (p=0.016) but not progression-free survival (p=0.116). Subgrouping patients by the prespecified first tertile (-0.23) of T-cell-inflamed GEP distribution yielded an objective response rate of 23% for GEPlow and 40% for GEPhigh (n=52). Tables 2 and 3 show the elaborated results.
Table 1. Baseline characteristics
Table 2. Associate of canonical signatures with clinical outcomes in the RNA-seq population adjusted for T-cell-inflamed GEP and IMDC
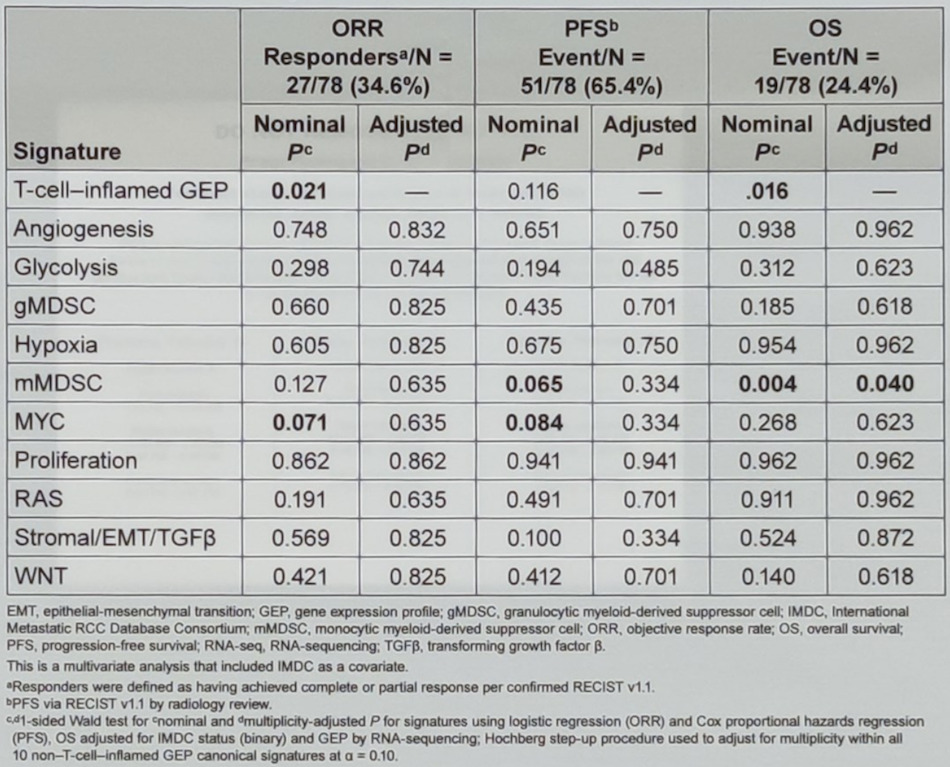
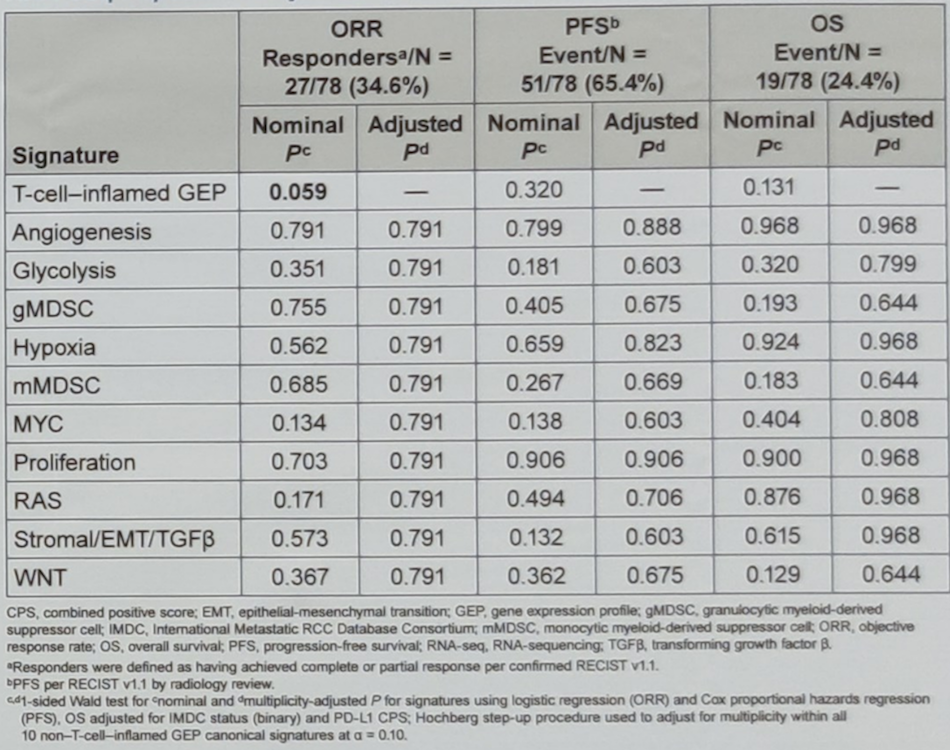
Table 3. Association of canonical signatures with clinical outcomes in the RNA-seq population adjusted for CPS and IMDC
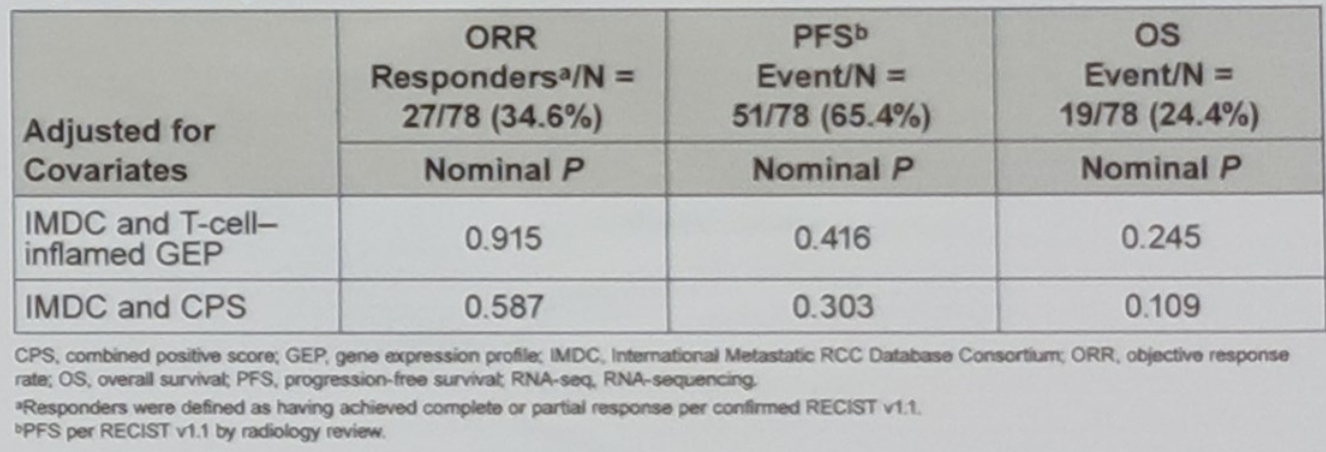
Data from TCGA shows a lower correlation between PD L1 and PD L2 in renal cell carcinoma samples than in samples of other tumor types. As a post hoc objective the authors evaluated mRNA PD-L2 levels to determine whether they offered explanatory value in patients with RCC. The testing results showed PD-L2 expression did not offer additional explanatory value beyond the T-cell inflamed GEP, consistent with the expectation that PD-L2 belongs to the larger T cell-inflammed process captured by the GEP (Table 4)
Table 4. Association of PD-L2 with clinical outcomes in the RNA-seq population adjusted for IMDC and T-cell-inflamed GEP or CPS
The authors concluded that RNA sequencing-based T-cell inflamed GEP was significantly associated with both objective response rate and overall survival in patients with clear cell renal cell carcinoma treated with first-line pembrolizumab monotherapy.
Aside from T-cell inflamed GEP signature, no other signatures showed a statistically significant association with the objective response rate, progression-free survival, or overall survival in all models analyzed.
Presented by: David McDermott, MD, Dana-Farber/Harvard Cancer Center, Boston, Massachusetts
Written by: Hanan Goldberg, MD, Urology Department, SUNY Upstate Medical University, Syracuse, New York, Twitter: @GoldbergHanan at the 2020 Genitourinary Cancers Symposium, ASCO GU #GU20, February 13-15, 2020, San Francisco, California


