TRITON2 is an international, multicenter, open-label, phase 2 study (CO-338-052; NCT02952534), which has aimed to determine how patients with mCRPC and evidence of a homologous recombination gene deficiency, respond to treatment with rucaparib, a poly(ADP-ribose) polymerase (PARP) inhibitor. In this ongoing study, evidence has accumulated showing that rucaparib has antitumor activity in BRCA1/2-deficient mCRPC tumors.5
In this study, the authors aimed to present associations of genomic characteristics and baseline clinical factors in 45 mCRPC patients with BRCA1/2 deficiency, who were enrolled in the ongoing TRITON2 study (enrollment cutoff: April 16, 2018; visit cutoff: June 29, 2018). In the TRITON2 study, eligible patients were screened for the presence of a deleterious germline or somatic alteration in BRCA1, BRCA2, or other prespecified DDR gene alteration.3,4 Central screening of tumor tissue or plasma was performed using next-generation sequencing assays by Foundation Medicine, Inc.6,7 Lastly, germline testing was performed for all patients using a Color Genomics assay.8
The demographic details of the enrolled patients are shown in Table 1. Most patients demonstrated frameshift alterations (49%, 22/45) or homozygous loss (27%, 12/45) of BRCA1/2 (Figure 1). Two-thirds of the patients (30/45) had a somatic, and 1/3 had a germline BRCA1/2 alteration. BRCA1/2 mutation zygosity was determined based on tissue profiling. A total of 36 patients submitted a tissue sample, 32 (89%) from prostate tumors, and 4 (11%) from metastases. Overall, 30 of 36 samples (83%) were sequenced successfully. BRCA1/2 alteration zygosity could be determined for 29 patients. Most patients (83%, 24/29) with known zygosity had a biallelic alteration.
Table 1 – Demographic details of the patients enrolled:
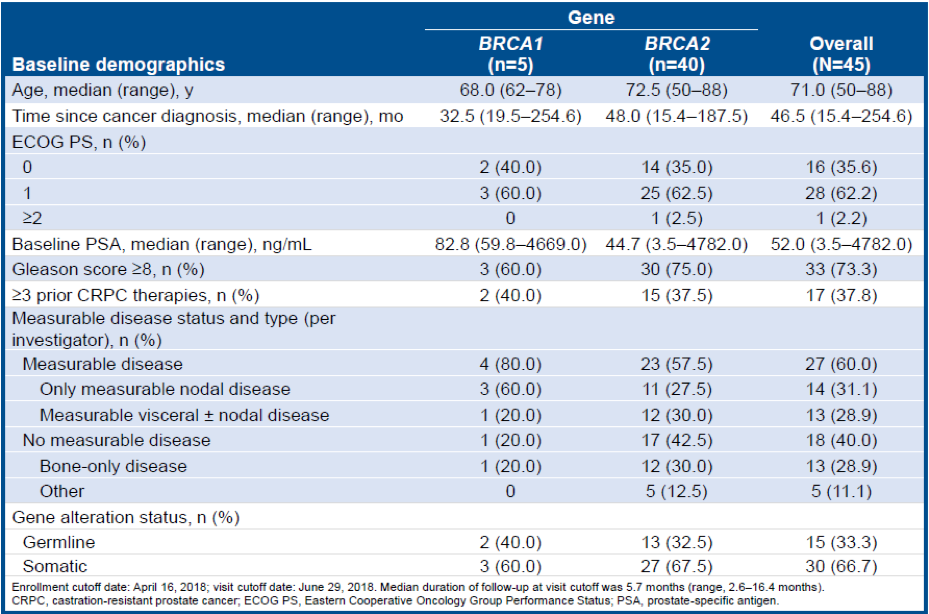
Figure 1 – BRCA 1 and 2 alteration types:
Patients were screened for deleterious DDR gene alterations using tissue or plasma samples. Plasma samples were taken at the time of progression on prior therapy and may reflect the current disease state more accurately than tissue samples. Circulating cell-free DNA (cfDNA) was purified from the plasma sample. A total of 89% (40/45) of the patients had plasma samples available for analysis. Most samples (98%,39/40) had sufficient cfDNA for successful sequencing. The plasma assay detected BRCA1/2 alterations in 94% (29/31) of patients.
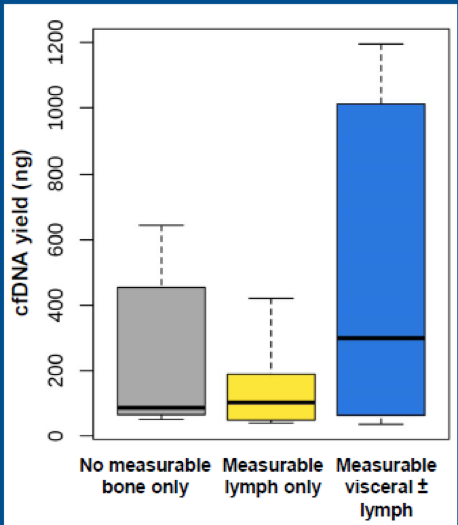
Plasma samples were collected within six weeks of the assessment of baseline clinical factors. The authors attempted to find associations between baseline cfDNA yield, cell-free tumor fraction, variant allele frequency, somatic/germline status, and baseline clinical factors. Figure 2 demonstrates that patients with visceral disease had the highest plasma cfDNA yield at baseline. No correlation was found between the baseline sum of target lesions and cfDNA yield (r=0.13, P=0.61). However, there was a trend toward patients with a larger sum of tumor lesions having higher tumor fraction
(r=0.46, P=0.07). Lastly, no correlation was found between baseline prostate-specific antigen (PSA) levels and cfDNA yield.
Figure 2 - cfDNA yield by disease type:
Of the patients with a known disease stage at diagnosis, 57% with a germline BRCA1/2 alteration was diagnosed with stage T3 or higher disease, compared to 42% of patients with a somatic BRCA1/2 alteration. Additionally, patients with a germline alteration were younger at the time of entering TRITON2 compared to patients with a somatic alteration. Also, the variant allele frequency was higher for germline than somatic alterations and was higher in younger patients (r=-0.66, P<0.01). Gleason score at diagnosis was higher in patients with higher allele frequency alterations.
In conclusion, this novel study reports that responses to rucaparib were observed in patients with germline or somatic BRCA1/2 alterations. Interestingly, compared to patients with a somatic BRCA1/2 alteration, patients with a germline BRCA1/2 alteration were diagnosed at a more advanced stage and were younger at the time of enrollment into TRITON2. More studies are needed in this specific population of patients to understand the various implications of BRCA 1 and 2 alterations will have on their treatment.
Presented by: Wassim Abida, MD, PhD, Oncologist, Memorial Sloan Kettering Cancer Center, New York, New York, United States
Written by: Hanan Goldberg, MD, Urologic Oncology Fellow (SUO), University of Toronto, Princess Margaret Cancer Centre, @GoldbergHanan at the 2019 ASCO Annual Meeting #ASCO19, May 31-June 4, 2019, Chicago, IL USA
References:
- Parker et al. "Cancer of the prostate: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up." Annals of Oncology. 2015. doi: 10.1093/annonc/mdv222.
- NCCN Clinical Practice Guidelines in Oncology. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf. Accessed October 10, 2018.
- Green et al. Presented at AACR; March 31, 2019; poster 727
- Chowdhury et al. "Genomic profiling of circulating tumour DNA (ctDNA) and tumour tissue for the evaluation of rucaparib in metastatic castration-resistant prostate cancer." Annals of Oncology. 2018.
- Abida et al. Ann Oncol. 2018;29(suppl 8):abst 793PD.
- Frampton et al. "Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing." Nature Biotechnology. 2013. doi: 10.1038/nbt.2696.
- Clark et al. "Analytical Validation of a Hybrid Capture-Based Next-Generation Sequencing Clinical Assay for Genomic Profiling of Cell-Free Circulating Tumor DNA."Journal of Molecular Diagnostics. 2018. doi: 10.1016/j.jmoldx.2018.05.004.
- Crawford et al. "Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients." Breast Cancer Research and Treatment. 2017. doi: 10.1007/s10549-017-4181-0.


