In PROpel, patients were enrolled irrespective of homologous recombination repair (HRR) gene mutation (HRRm) status. However, the authors examined key outcomes stratified by HRRm status. In general, tumor-based testing is considered the gold standard, however, there are limitations in obtaining sufficient quality tissue for analysis, thus the authors also examined ctDNA.
Among all enrolled patients, more than 98% had available archival tumor tissue and blood samples at baseline. These were analysed using FoundationOne®CDx and FoundationOneLiquid®CDx molecular diagnostic tests for tumor tissue and ctDNA, respectively. HRRm status (HRRm, non-HRRm or HRRm unknown) was assigned by individual test result and using an aggregate of the results from both tests. When considered in aggregate, patients were classified as HRRm if an HRRm was detected by either test; non-HRRm if no HRRm was detected by either test; or HRRm unknown if they did not have a valid HRRm test result.
Among the 796 randomized patients in the trial, 782 had tumor tissue samples and 794 had blood samples available for testing. Among those patients with tumor tissue samples, 535 (68%) were successfully assessed for HRRm status by tumor tissue testings, while 734 (92%) of patients with blood samples were successfully assessed by ctDNA test.
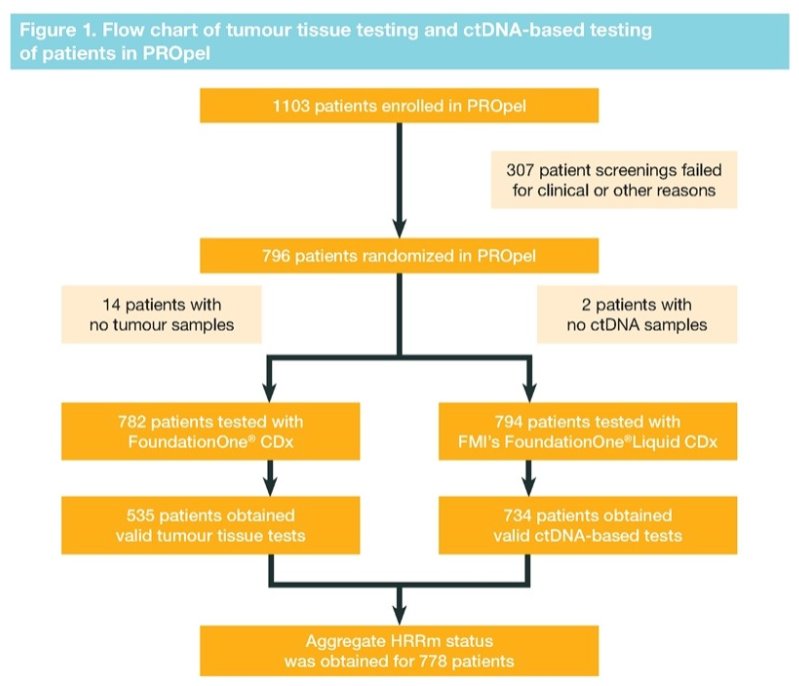
Aggregate HRRm status was obtained for 778 (98%) patients of whom 226 were classified as HRRm, 552 as non-HRRm and 18 as HRRm unknown. Using tumor tissue as a reference, positive-percent agreement was 80%, negative-percent agreement was 87%, overall-percent agreement was 85% and the positive and negative predictive values were 64% and 94%, respectively.
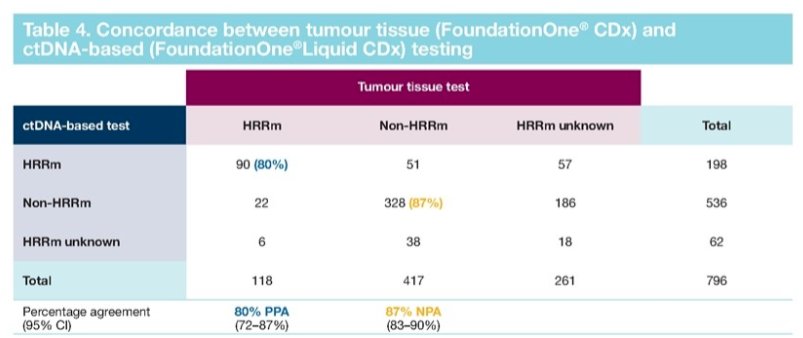
In the HRRm by ctDNA subgroup, 51 patients were non-HRRm by tumor tissue. The negative predictive value suggests that in the non-HRRm by ctDNA subgroup, 174/186 (94%) patients would also be non-HRRm by tumor tissue test. Therefore, there were approximately 12 potential false negative results by ctDNA test in the total non-HRRm aggregate population (2% of 552 patients).
In conclusion, Dr. Armstrong noted that there was good agreement between matched tumor tissue and ctDNA test results in PROpel. Using aggregate test results, it was possible to maximize the number of patients with known HRRm status while controlling the number of potential false negative results, thus facilitating robust analysis of HRRm status.
Presented by: Andrew J. Armstrong, MD, MSc, Medical Oncologist, Duke Health, Durham, NC
Written by: Christopher J.D. Wallis, University of Toronto, Twitter: @WallisCJD during the 2022 European Society of Medical Oncology (ESMO) Annual Hybrid Meeting, Paris, FR, Fri, Sept 9 – Tues, Sept 13, 2022.


