(UroToday.com) The ANZUP 2021 virtual annual scientific meeting included a presentation by Dr. Eli Van Allen discussing the convergence of machine learning and genomics for precision cancer medicine in genitourinary cancers and beyond. As both a medical oncologist and a clinician he utilizes computational biology to assess canceromic biology for the improvement of cancer outcomes. Dr. Van Allen notes that for the last 15-20 years, there has been expanding data at the point of care, including the cost of sequencing genomes substantially decreasing and the data points available per patient increasing. This includes: whole exomes, genomes, transcriptomes, epigenomics, histopathology and unstructured electronic health record data, as summarized in the following figure:
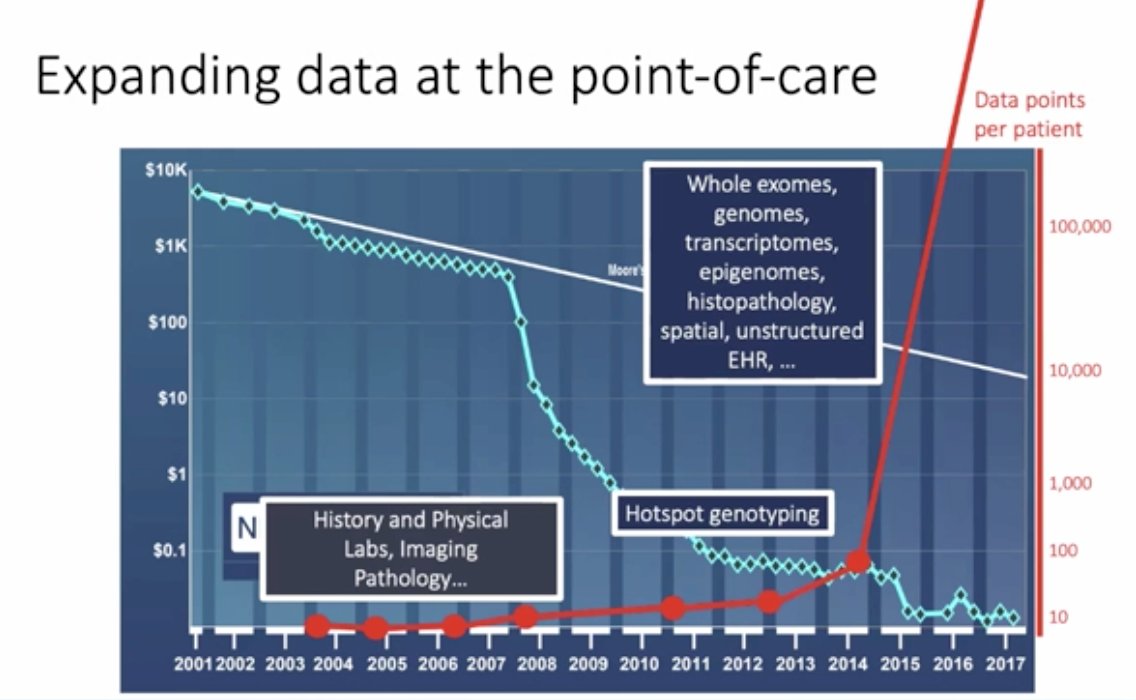
Based on this premise, Dr. Van Allen emphasizes that this has led to the primary question that his lab is attempting to answer: Can expanded molecular profiling at the point-of-care guide individualized treatment in oncology? Dr. Van Allen notes that in 2013 the clinical interpretation of cancer genomics included Precision Heuristics for Interpreting the Alteration Landscape (PHIAL), defined as a heuristic-based clinical interpretation algorithm that sorted somatic variants from whole exomes by clinical and biological relevance. Their group subsequently built a database based on the PHIAL algorithm called the Tumor Alterations Relevant for Genomics-Driven Therapy (TARGET), which is a database of genes that may have therapeutic, prognostic, and diagnostic implications for patients with cancer.
Dr. Van Allen notes that there are several challenges to clinical interpretation of genomic data in 2021. First, technology in the clinic is evolving, moving from DNA only to expanded DNA (bulk RNA) to expanded DNA including bulk RNA, single cell, immune assays, and multiplex immunohistochemistry. Second, our knowledge keeps evolving, whereby the data based on the TARGET database (built in the 2010’s) including only 50% relevant information in 2021. The third challenge is that clinicians need help, given that computation cancer biologists were providing reports for clinicians with esoteric jargon that was creating confusion and frustration when applying this information to patient-centered care.
There are several ways that computational oncologists can help clinicians. First, interpreting algorithms for evolving technology at the point of care, and annotating and evaluating second-order genomic relationships:
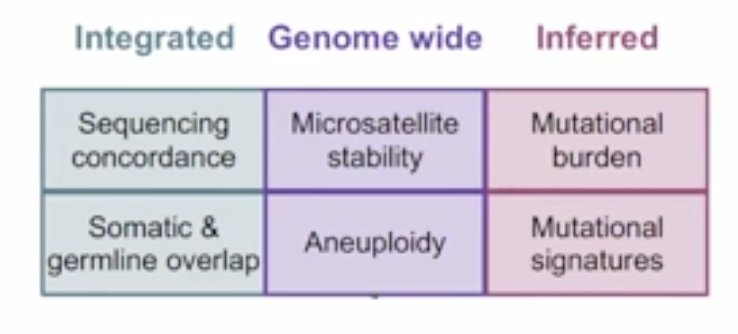
Each of the above boxes can apply a deep learning germline variant model for cancer patients, proving that it outperforms the current standard of care for finding these actionable variants in the first place. Second, additional preclinical value can be added to clinical interpretation. By creating a match-making program, computational oncologists are able to take a patient’s molecular profile and find which cell lines are most similar to that patient, link the drugs that make these cell lines disappear, and bring them into the patient’s actionability report. Third, Dr. Van Allen and his team have been able to take a standard report and develop an enhanced, web-based report that allows easier actionability for patient care.
What Dr. Van Allen and his team have been able to do over the last several years is take all of the information they’ve generated and redo the entire concept of clinical interpretation of cancer genomics, resulting in the Molecular Oncology Almanac (www.moalmanac.org). This includes taking a patient’s molecular data (DNA and RNA), matches their first and second order events, does further match-making between patients and cell lines for similarity, and subsequently generates a report.
Dr. Van Allen notes that despite the progress being made, there is still a long way to go, for example, in advanced prostate cancer and developing a molecular signature, predicting which patients will have advanced disease. In early work from his group, Dr. Van Allen notes that they used 1,053 prostate cancer samples to attempt to develop a molecular signature (based on TP53, PTEN, and AR) for predicting primary versus castration-resistant prostate cancer classification; however, they noted that ultimately the model was not very good. Based on these poor results, they realized that new methodologies must be developed for biological discovery, therapeutic target identification, and clinical patient risk stratification. As such, over the last several years, they have attempted to integrate biology and machine learning for molecular discovery, resulting in P-NET (a biological informed neural network) [1]:
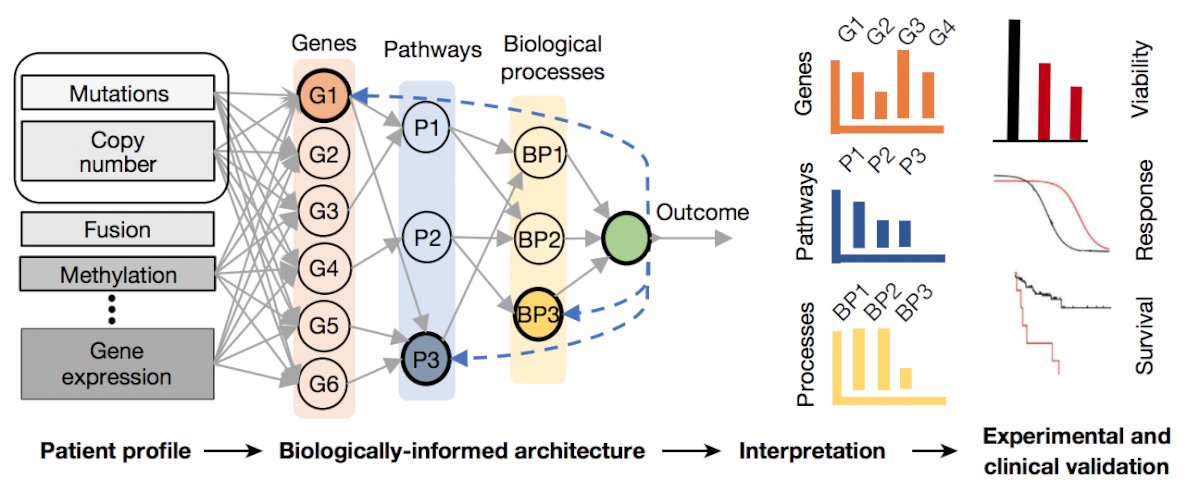
This study demonstrated that P-NET can predict cancer state using molecular data with a performance that is superior to other modelling approaches. Moreover, the biological interpretability within P-NET revealed established and novel molecularly altered candidates, such as MDM4 and FGFR1, which were implicated in predicting advanced disease and validated in vitro.
There are two ways for generating more data to feed into these networks. First, capturing tumors and delineating immune and stromal programs, including data from pre-treatment biopsy + blood (bulk whole exome sequencing/whole transcriptome sequencing, scRNASeq, models, and liquid whole exome sequencing), on-therapy biopsies + blood (bulk whole exome sequencing, scRNASeq, and liquid whole exome sequencing), and resistance biopsy + blood (bulk whole exome sequencing/whole transcriptome sequencing, scRNASeq, models, and liquid whole exome sequencing). Second, The Metastatic Prostate Cancer Project is integrating data sets for discovery by taking research to the patient’s doorstep. Once patients opt into this collaborative, several kits (saliva, etc) are then sent to the patient (which may be over treatment time to assess the possibilities of resistance) in order to generate data. So far, over 1,000 men with metastatic prostate cancer have joined The Metastatic Prostate Cancer Project since the launch in January 2018.
Presented by: Eliezer (Eli) Van Allen, MD, Medical Oncology, Dana-Farber Cancer Institute, Boston, MA
Written by: Zachary Klaassen, MD, MSc – Urologic Oncologist, Assistant Professor of Urology, Georgia Cancer Center, Augusta University/Medical College of Georgia, @zklaassen_md on Twitter during the 2021 Australian and New Zealand Urogenital and Prostate (ANZUP) Cancer Trials Group Annual Scientific Meeting (ASM), Sunday, Oct 17 – Monday, Oct 18, 2021.
References:
- Elmarakeby HA, Hwang J, Arafeh R, et al. Biologically informed deep neural network for prostate cancer discovery. Nature. 2021;598:348-352.


